I think this example in C++ should work. Unfortunately I don't know Python DistanceTranform but I think you can use this tutorial in C++ and this one in python to translate it in python. SparseMatrix is only for fun. you don't need it result (in Mat result) is saved in an yml file.
int main(int argc, char* argv[])
{
Mat img=imread("14878460214049233.jpg",IMREAD_GRAYSCALE);
imshow("test",img);
threshold(img,img,200,255,CV_THRESH_BINARY); // to delete some noise
imshow("test", img);
Mat labels;
connectedComponents(img,labels,8,CV_16U);
Mat result(img.size(),CV_32FC1,Scalar::all(0));
for (int i = 0; i <= 1; i++)
{
Mat mask1 = labels == 1+i;
Mat mask2 = labels == 1+(1-i);
Mat masknot;
bitwise_not(mask1,masknot);
imshow("masknot", masknot);
Mat dist;
distanceTransform(masknot,dist, DIST_L2,5,CV_8U);
imshow("distance float", dist/255);
dist.copyTo(result,mask2);
}
imshow("distance 1",result);
FileStorage fs("distCtr.yml",FileStorage::WRITE);
fs<<"Image"<<result;
fs.release();
waitKey();
SparseMat ms(result);
SparseMatConstIterator_<float> it = ms.begin<float>(),it_end = ms.end<float>();
Mat lig(result.rows,1,CV_8U,Scalar::all(0));
for (; it != it_end; it ++)
{
// print element indices and the element value
const SparseMat::Node* n = it.node();
if (lig.at<uchar>(n->idx[0])==0)
{
cout<< "("<<n->idx[0]<<","<<n->idx[1]<<") = " <<it.value<float>()<<"\t";
lig.at<uchar>(n->idx[0])=1;
}
}
return 0;
}
 (/upfiles/14878459581269807.jpg)
(/upfiles/14878459581269807.jpg) 
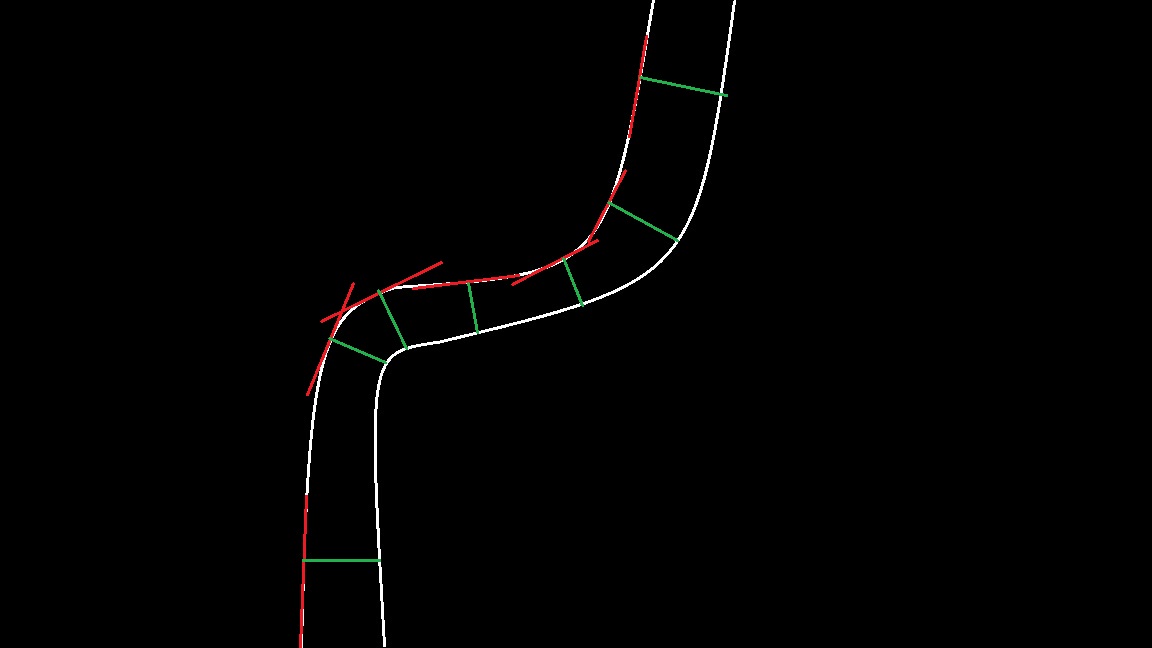
what about cv::distanceTransform (like suggested by @sturkmen )
How would I implement the cv::distanceTransform? Cant get it to work :/
Actually, I don't think distance transform is what he wants. That's just the shortest distance, not the perpendicular distance.
I can't think of a particularly fast way of doing perpendicular distances. If you can accept using just the distance to the closest point on the other line, the distance transform is great.